Google Summer of Code Final Work Product
SunTzunami
Published: 08/23/2021
Google Summer of Code Final Work Product
Proposed Objectives
- Ribbon Representation
- Molecular Surface Representation
- Stretch Goals:
- Stick Representation
- Ball and stick Representation
- Wire Representation
- Pipes and Planks Representation
- Sphere Representation
Objectives Completed
- Ribbon Representation
Ribbon diagrams, also known as Richardson diagrams, are 3D schematic representations of protein structure. Ribbon diagrams are generated by interpolating a smooth curve through the polypeptide backbone. α-helices are shown as coiled ribbons. β-strands as sheets, and non-repetitive coils or loops as lines or thin strips. It was implemented by using vtkProteinRibbonFilter. Generating a vtkPolyData of appropriate format required by vtkProteinRibbonFilter was initially unclear due to lack of examples. I was able to understand what kind of output the filter required after a meeting with mentors. Tests were added and a demo was created.
Pull Requests:
- Ball and Stick Representation
The ball-and-stick model is a molecular model of a chemical substance which displays both the three-dimensional position of the atoms and the bonds between them. The atoms are typically represented by spheres, connected by tubes which represent the bonds. It was created by using vtkOpenGLMoleculeMapper. Added vtkSimpleBondPerceiver for detection of bonds. Tests were added and a demo was created.
Pull Requests:
- Stick Representation
Stick model is a special case of Ball and Stick model where atomic radius of all molecules is set equal to the radius of tubes used to create bonds. It was created by using vtkOpenGLMoleculeMapper. Tests were added and a demo was created.
Pull Requests:
- Sphere Representation
In chemistry, a space-filling model, also known as a calotte or sphere model, is a type of three-dimensional (3D) molecular model where the atoms are represented by spheres whose radii are proportional to the radii of the atoms. It was created by using vtkOpenGLMoleculeMapper. Tests were added and a demo was created.
Pull Requests:
Objectives in Progress
- Molecular Surfaces
There are three types of molecular Surfaces:
- Van der Waals
- Solvent Accessible
- Solvent Excluded
Currently the first two molecular surfaces i.e. Van der Waals and Solvent Accessible are implemented. The code is based on the paper "Generating Triangulated Macromolecular Surfaces by Euclidean Distance Transform" by Dong Xu and Yang Zhang.
Pull Requests:
Other Objectives
- 2D Animated Surfaces
This was a simple demonstration that animated Two-Dimensional (2D) functions using FURY. Created a grid of x-y coordinates and mapped the heights (z-values) to the corresponding x, y coordinates to generate the surfaces. Used colormaps to color the surfaces.
Pull Requests:
- Updated miscellaneous animations
- Updated the demo of helical motion to stop using multiple line actors as discussed in the meeting.
- Updated the demo of brownian motion to make it more scientifically useful (removed unnecessary rotation of camera during animation and box actor).
- Display simulation data for brownian motion and helical motion animations (number of simulated steps for brownian motion and velocity of the particle for helical motion).
- Created utility functions to make the code understandable and used these in emwave, helical and brownian animations.
Pull Requests:
- GSoC weekly Blogs
Weekly blogs were added for FURY's Website.
Pull Requests:
Timeline
Detailed weekly tasks and work done can be found here.
View Blog Post
Weekly Check-in #11
SunTzunami
Published: 08/16/2021
Tenth/Final week of coding
Welcome to the eleventh and final weekly check-in. This was the tenth and final week of coding.
The summer went by swiftly and the coding period has come to an end. It was a great learning experience. Two crucial technical skills that I enhanced over this summer:
- Ability to understand documentation of different packages
- Ability to implement algorithms from scientific papers
I was able to evolve my problem solving approach thanks to the challenges I faced during the coding period.
My mentors, core team and the FURY community were extremely supportive and helped me whenever I got stuck. I'd like to thank Nasim and Elefth for helping me with the theoretical aspects of the project and helping me understand the research papers and underlying math. I'd like to thank Serge for his code reviews which helped me improve my code quality and helped me make my code more pythonic. I'd like to thank Javier for helping me with the Ribbon representation (which was something that I had been stuck on for a while). Javier also showed me the projects which he was working on which were truly fascinating and also inspired me to work on projects of my own.
One crucial learning outcome over the course of coding period:
Ability to break down problems into smaller pieces (decomposition). Most of the times, I tried to implement everything in one go. This is not a good approach and gets problematic when what you're trying to implement is complicated. Segmenting a problem's solution into steps is the way to go. A time when I employed this approach of segmenting the implementation into several steps during GSoC was when writing the code for the molecular surfaces which involved the following steps:
- Creating a grid of a specific resolution
- Finding the ids of voxels composing the molecular surface
- Implementing marching cubes or some other isosurface generation algorithm
- Smoothen the isosurface obtained in previous step
I'll be sharing my progress for the final week of coding now. The github repo of FURY can be found here.
What did you do this week?
- Implemented this paper (PR created: PR #492) to generate Van der Waals surface and solvent-accessible surface. It was a good learning experience because the first time I read the paper, I didn't understand the underlying math, it all seemed alien to me. I had to read it many times, read up the algorithms used and understand the terminologies. I had a meeting with the mentors to understand a bit of the theory which proved to be quite fruitful as I understood how to go about making the space-filling model. This blog was kinda helpful in understanding how to use
vtkMarchingCubes with numpy arrays. One of the earliest SAS rendering looked like this (this implementation was not strictly according to the paper):
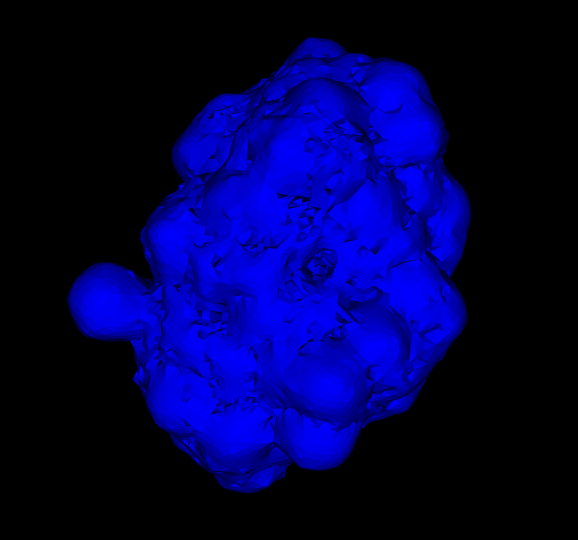
Current implementation (this implementation was according to the paper):

grid dimenstions = 256 × 256 × 256, used smoothing algorithms recommended by vtk
- I also understood how to go about rendering volumes. I think that the ability to render volumes with FURY will be a cool capability and I'll discuss my implementation and request the mentors for feedback and ideas in the weekly meeting. Example of volume rendering:

What is coming up next week?
I'll try to get PR #452 merged. Documentation work to be done as GSoC coding period has come to an end.
Did you get stuck anywhere?
The paper I was trying to implement seemed quite intimidating at first. Understanding it and the underlying math took a few days. Implementing it in python was also a challenge as usage of vtkMarchingCubes with numpy arrays is not that well documented. All in all, it was a nice learning experience.
Parting Words
In retrospect, it wasn't always smooth sailing as I used to get stuck quite often (sometimes for weeks) while working on certain representations (VTK documentation wasn't quite helpful and didn't have many python examples). But I didn't give up and read hundreds (if not thousands) of blogs and many research papers to find the solutions to the problems that I encountered (I read over 600 topics on the VTK forum). The key to breaking an impasse is to be unrelenting in the pursuit of answers. Also, don't hesitate to seek help as most of the times the impasse that one faces is something that others have encountered before and they might offer some insight that can help. This pursuit gets quite frustrating at times but the satisfaction one feels after getting the answers makes this struggle worthwhile (quitting ain't an option!).
The coding period has ended and my GSoC journey too is nearing its end but I believe that I'm just getting started with Open Source and I can't wait to work on the ideas I developed over the course of this summer. I have some exciting project ideas for FURY (physics and animation based) that I hope to work on.
To the Reader,
Good luck with your future endeavours and goals.
Never give up.
Au Revoir :)

View Blog Post
Weekly Check-in #10
SunTzunami
Published: 08/09/2021
Ninth week of coding
Welcome to the tenth weekly check-in. I'll be sharing my progress for the ninth week of coding.
What did you do this week?
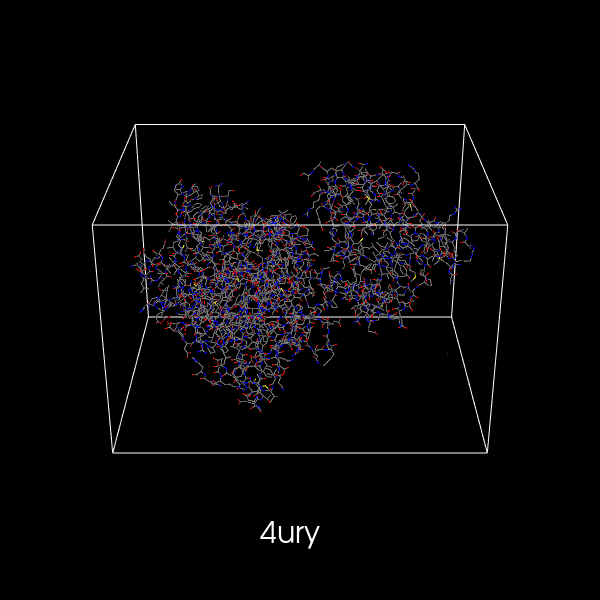
- Updated PR #452:
- Made ribbon representation faster.
- Added an actor to display bounding box around the molecule.
Preview:
- Made a tutorial which showcases the abilities of
molecular module (will create a PR after molecular module is merged).
- I'm trying to implement a native implementation of molecular surfaces in FURY. Currently searching for recent research papers to find good algorithms to generate the molecular surfaces.(the ones I'd collected in the research period were archaic and rather time consuming). The papers that I've read so far seem a tad bit intimidating as I've never done math related to this domain yet. Implementing them will be a good learning experience I reckon.
What is coming up next week?
- Try to create a native implementation of molecular surface.
- Small fixes to PR #362, PR #462.
Did you get stuck anywhere?
No.
View Blog Post
Weekly Check-in #9
SunTzunami
Published: 08/02/2021
Eighth week of coding
Welcome to the ninth weekly check-in. I'll be sharing my progress for the eighth week of coding.
What did you do this week?
- Updated PR #452: Had an extra meeting with the mentors in which we fine-tuned the
molecular module and optimized it to make it more pythonic.
- I was able to generate vertices and triangles for Solvent Excluded Surfaces (a type of molecular surface representation popularly called SES) by using a bioconda package called msms.
It's based on this paper by
Michel F. Sanner, Arthur J. Olson & Jean-Claude Spehner. It was then rendered via
surface actor.
Preview (SES surface generated by using msms):

- Added my GSoC blogs to the FURY blogs directory. (PR #475)
What is coming up next week?
- Research about recent papers having good (fast) algorithms to create the molecular surfaces.
- Create tutorials to explain how to use
molecular module.
Other goals will be decided in the meeting with mentors.
Did you get stuck anywhere?
No.
View Blog Post
Weekly Check-in #8
SunTzunami
Published: 07/26/2021
Seventh week of coding
Welcome to the eighth weekly check-in. I'll be sharing my progress for the seventh week of coding.
What did you do this week?
- Updated PR #452:
- Added Ribbon representation to the
molecular module.
- Added tests for all functions and classes of
molecular module. Writing tests is kinda fascinating.
- Updated PR #462: Addressed the reviews of team members and mentors, added more new features.
Preview:

- Updated PR #362: Addressed the feedbacks of team members and mentors.
What is coming up next week?
- Work more on
molecular module, meeting with mentors and core team on thursday to optimize the module and merge PR #452.
- Work upon molecular surface representation.
Did you get stuck anywhere?
No.
View Blog Post






